InTraSeq™ Single Cell Analysis Overview
This page provides an in-depth overview of the InTraSeq technology, how it can empower your single-cell analysis research, and product offerings.
Use the links below to jump to your topics of interest.
Seq What You’ve Been Missing from Your Single-Cell Analysis
InTraSeq Single Cell Analysis Features & Advantages
Optimize Your Single-Cell Analysis Research
Seq What You’ve Been Missing from Your Singe-Cell Analysis
Analysis of isolated CD4+ and CD8+ cells using InTraSeq intracellular and signaling pathway antibodies to identify cellular states that would be difficult to study using RNA alone. The InTraSeq assay offers additional insights into cellular subpopulations, by measuring PTMs and intracellular protein levels in single cells, enabling a deeper understanding of disease development mechanisms.
Intracellular protein and Transcriptomic Sequencing (InTraSeq) is a technology for single-cell analysis developed by CST that identifies signaling pathways and reveals molecular mechanisms in disease development in a single experiment. It enables simultaneous detection of RNA as well as both intracellular and surface proteins in thousands of cells, allowing researchers to investigate signaling pathways with the transcriptome—all at a single-cell resolution.
InTraSeq facilitates deeper understanding of cell biology by:
- Uncovering biological insights not revealed with traditional single-cell RNA sequencing (scRNA-seq) methods
- Highlighting the dynamics between transcription and translation in single-cell datasets
- Dissecting cell signaling mechanisms by quantifying post-translational modifications at the single-cell level
InTraSeq 3’ technology is developed and validated by CST, using the 10x Genomics Chromium Single Cell 3' Reagent Kits with Feature Barcoding technology.
Most currently available single-cell analysis technologies are limited to phenotyping a sample because they only identify RNA and, occasionally, surface marker expression levels. Additionally, other technologies are not able to determine and measure important signaling cascades activated during a disease state or by pharmacologic perturbation. By combining transcriptomic, protein, and post-translational modifications quantification into a single assay, InTraSeq technology goes beyond phenotyping and dissects the cellular mechanisms that distinguish cellular differences—all in one experiment. With InTraSeq technology, scientists can readily explore multiple signaling pathways along with genetic expression changes to efficiently identify key molecular mechanisms underlying physiologic responses and discern downstream protein modifications.
Single-Cell Analysis (SCA) for Intracellular Proteins and Post-Translational Modifications
Single-cell RNA sequencing (scRNA-seq) is commonly utilized to determine cellular responses to pharmacologic and environmental perturbations. However, protein expression levels often do not directly correlate with RNA expression, leaving a gap in the understanding of complex physiologic systems. Determining how single-cell protein expression and post-translational modifications differ from single-cell gene expression can offer functional insight into pathway activation that cannot be obtained from scRNA-seq alone.
Post-translational modifications (PTMs) can alter protein activity, longevity, and expression levels. Understanding the interplay between genetic expression and protein activity is crucial to determine the underlying cellular heterogeneity and functional state of complex biological systems. The InTraSeq 3' Conjugate Antibody Cocktail is formulated with 31 of our most popular antibodies that bind to both human and mouse targets, including PTMs. Scientists can quantify dozens of critical signaling proteins in a single assay with ease and without bias.
Co-Quantification of Single-Cell PTMs and mRNA
Many cells can exhibit similar levels of RNA expression, but some may differ in which proteins are ultimately expressed and subsequently modified to regulate activity after pharmacologic or environmental perturbations. Co-quantification of RNA, protein, and post-translational modifications facilitates a more thorough understanding of complex physiologic interactions that RNA expression alone cannot elucidate.
RNA Does Not Necessarily Correlate with Protein Levels
InTraSeq Single Cell Analysis Features & Advantages
Reduced Hands-On Time with a Streamlined Workflow
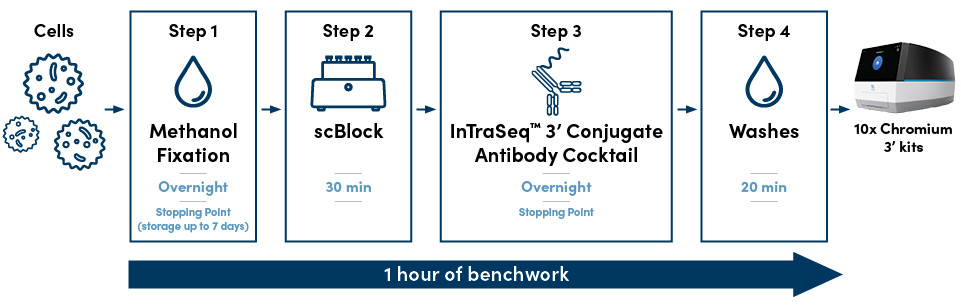
A Straightforward, Four-Step Protocol
The InTraSeq protocol requires approximately one hour of hands-on bench time and includes multiple stopping points, providing maximum time flexibility when performing single-cell experiments. Samples can be stored for up to seven days after the fixation step without compromising RNA integrity. Unlike currently available single-cell analysis protocols, the convenience offered by InTraSeq eliminates the need to coordinate staff and equipment with long single-day protocols. The ability to store samples for longer periods of time can also facilitate high-throughput applications by allowing bulk sample and single-cell processing protocols to be performed on sequential days without compromising quality.
The straightforward immunostaining protocol includes the following steps:
Step 1: Fix the cells overnight.
- ~5 min hands-on benchwork
- Cells can be stored in the freezer for up to seven days.
Step 2: Incubate with scBlock.
- ~10 min hands-on benchwork
- This step is optimized to obtain a high-quality single-cell readout of both RNA and proteins.
Step 3: Add CST® InTraSeq™ 3’ Conjugate Antibody Cocktail, and incubate overnight.
- ~5 min hands-on benchwork
Step 4: Wash the cells.
- ~20 min hands-on benchwork
- At this point, the cells are ready for a single-cell 10x Genomics 3’ experiment.
Investigate Intracellular Protein Signaling while Guaranteeing Robust RNA Signal
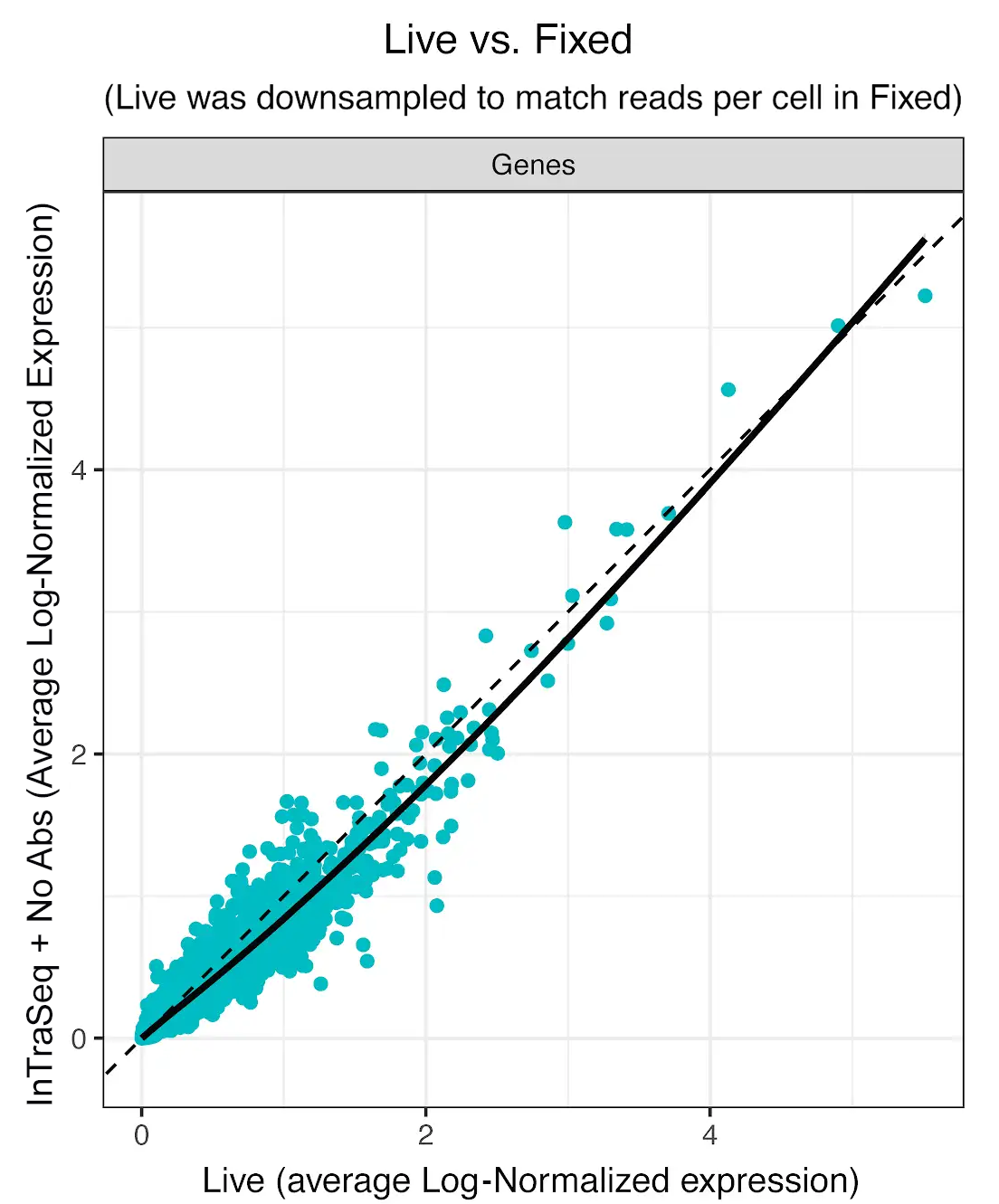
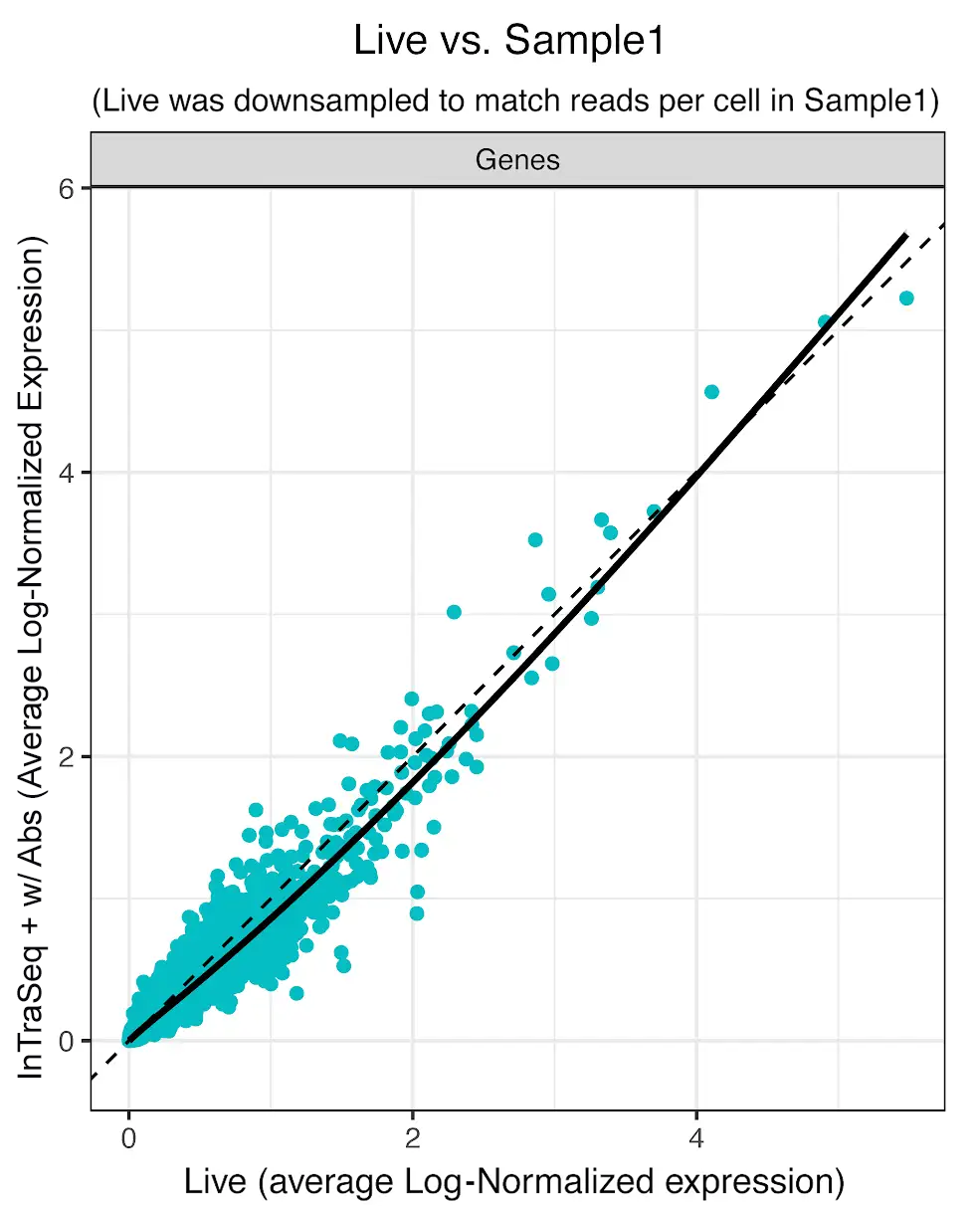
Preserving RNA integrity in single-cell multiomic experiments has been a persistent and significant challenge. Many single-cell assays utilize harsh chemicals to disrupt the membrane, resulting in RNA degradation and loss. InTraSeq reagents have been rigorously optimized to gently permeabilize the cell and nuclear membranes. This allows the conjugated antibodies to enter the cell and bind to their targets while maintaining RNA levels throughout the assay. This enables accurate quantitation of both RNA and protein expression.
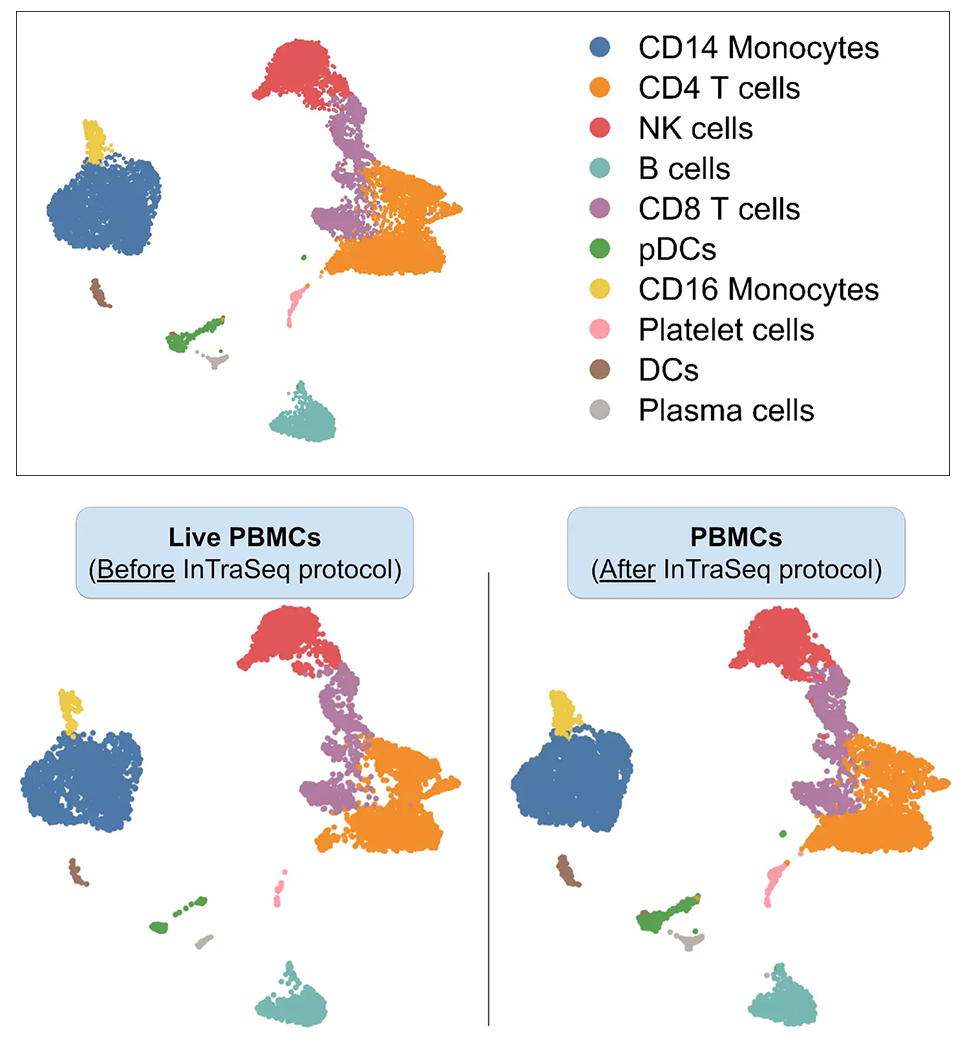
InTraSeq Preserves the Transcript Abundance in the Sample Compared to Live Cell Control
| Live PBMCs | InTraSeq™ PBMCs |
|---|---|---|
Relative Median Genes/Cell (matched reads/cell) | 100% | 86.2% |
Fraction Reads in Cells | 94.9% | 85% |
Antibody Reads in Cells | N/A | 60% |
Reads Mapped Confidently to Genome | 89.9% | 85.5% |
Average gene expression was quantified in live and InTraSeq prepared samples of PMBCs. InTraSeq reagents preserve the genetic profile of live cells in difficult samples such as PBMCs, with minimal loss of mRNA.
Identify Hard-To-Detect Cells with Unbiased Depth-of-Coverage
The power of InTraSeq Single Cell Analysis technology lies in the ability to detect and quantify both intracellular and surface proteins in conjunction with single-cell gene expression to differentiate heterogeneous cell populations. In contrast to CITE-Seq, which only detects cell surface proteins, InTraSeq provides scientists with tools to study complex intracellular signaling pathways without bias.
InTraSeq Method Preserves Cellular Heterogeneity
Explore Multiple Molecular Mechanisms In One Experiment
Combined with CST expertise in intracellular signaling and post-translational modification antibodies, the InTraSeq technology enables researchers to study and quantify intricate intracellular, nuclear, and surface protein interactions together with gene expression. Simultaneously measuring transcriptomics changes and protein expression, including post-translational modifications, generates more comprehensive data sets, enriching the knowledge of complex signaling systems in a way not possible with other assays and deepening our understanding of disease mechanisms.
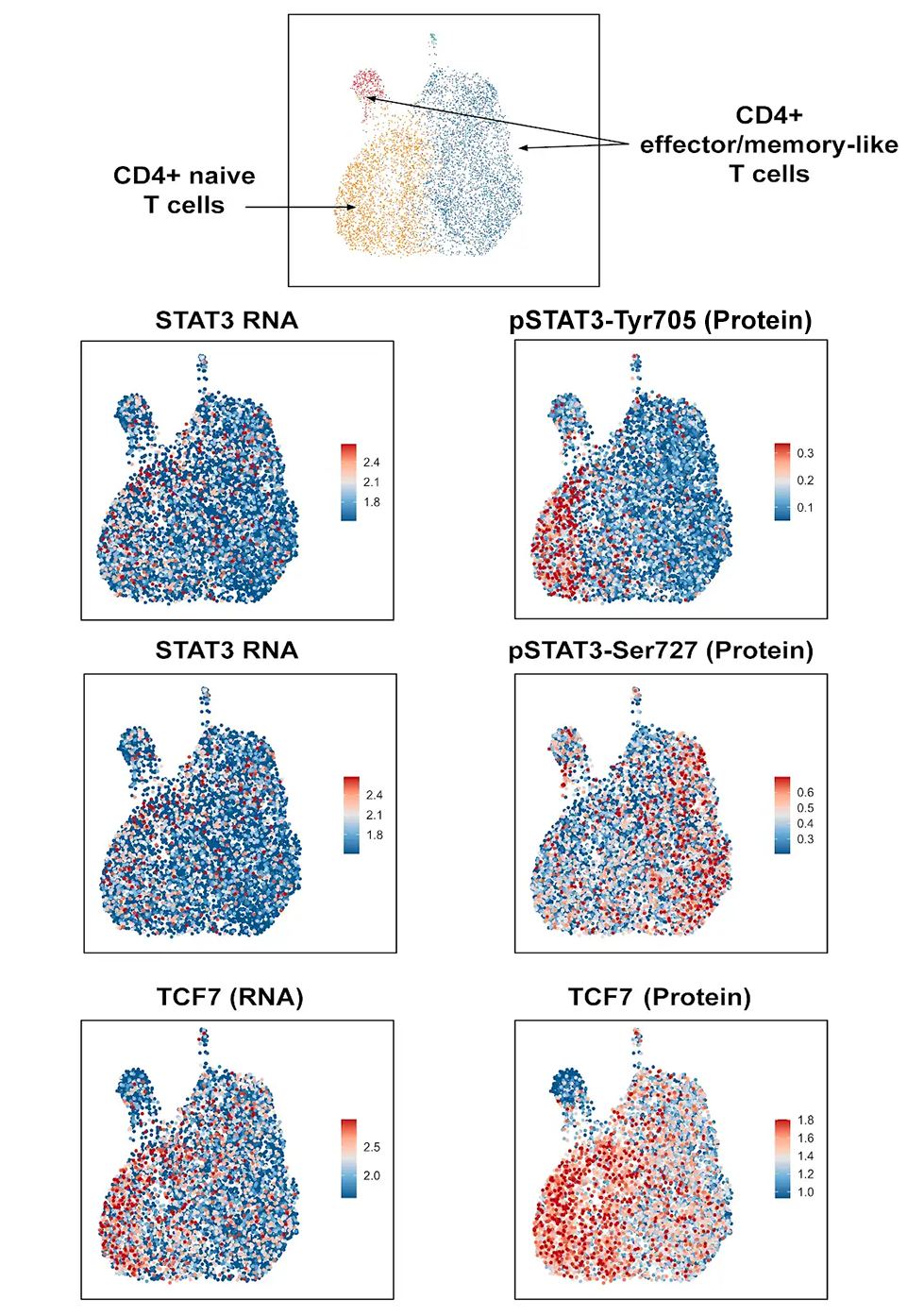
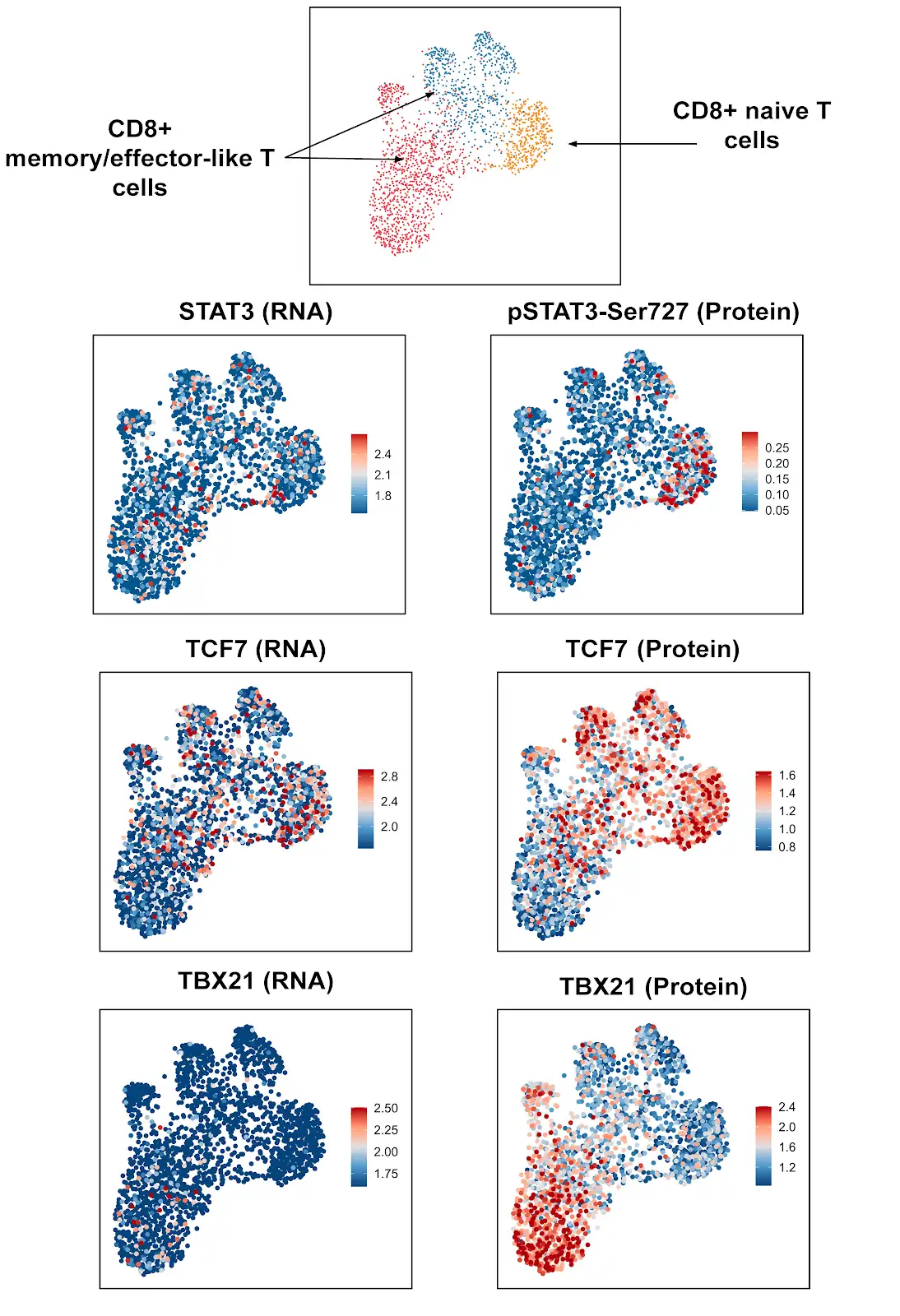
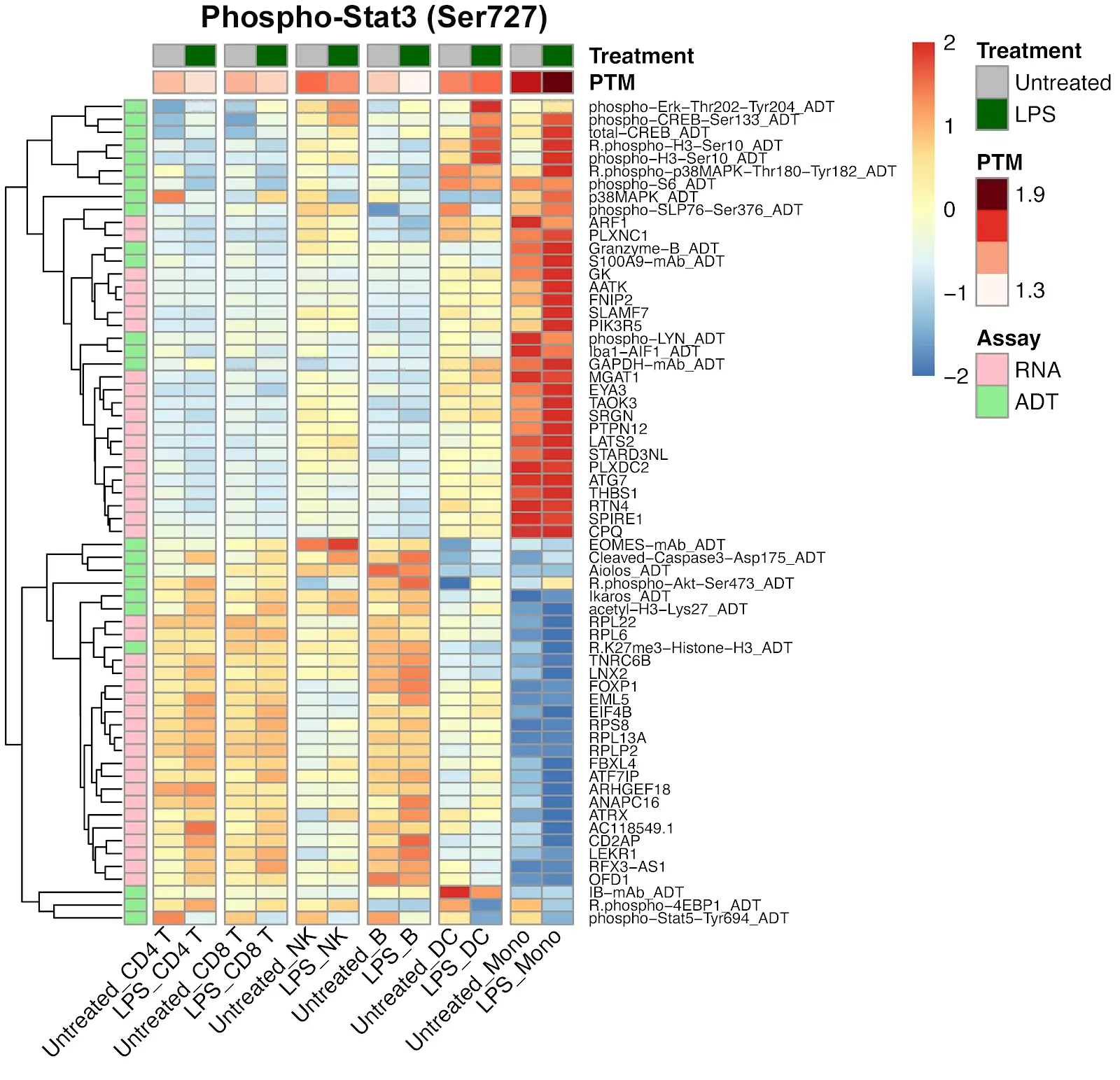
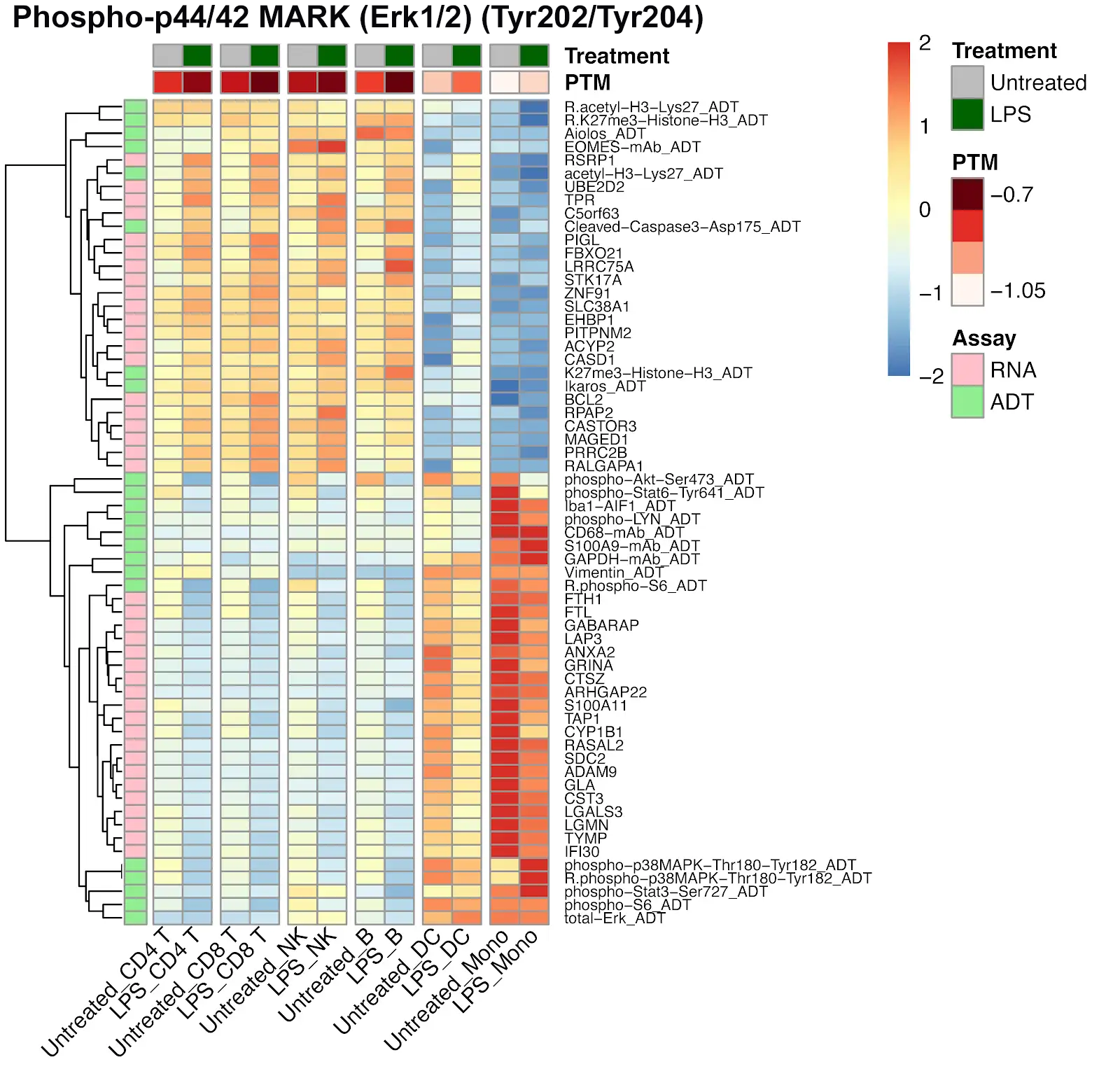
Identifying Cell States within CD4+ and CD8+ Cells Using Single Cell Intracellular Protein Readout
Analysis of isolated CD4+ and CD8+ cells using InTraSeq intracellular and signaling pathway antibodies to identify cellular states that would be difficult to study using RNA alone. The InTraSeq assay offers additional insights into cellular subpopulations, by measuring PTMs and intracellular protein levels in single cells, which can enable a deeper understanding of disease development mechanisms.
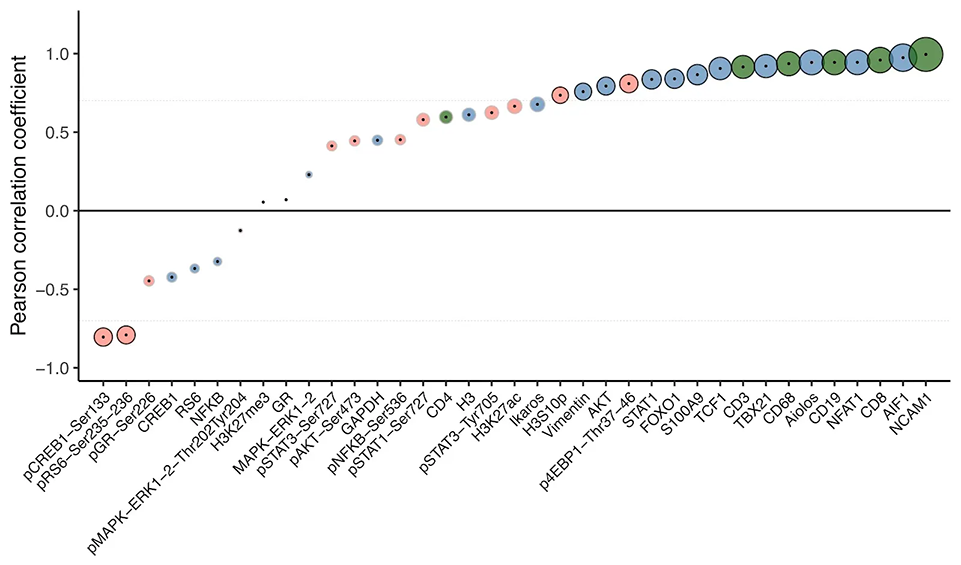
Of the 31 validated antibodies in InTraSeq 3' Conjugate Antibody Cocktail, 2 target surface proteins, and the other 29 target intracellular proteins, including PTMs. The antibody cocktail is designed for unbiased quantitation of proteins along signaling pathways commonly studied in immunology and cancer research, providing accurate and reliable data sets.
PBMCs were stimulated with LPS and processed using the InTraSeq™ Assay Kit, RNA, total protein, and PTMs were quantified with scRNA-seq and the InTraSeq™ 3' Conjugate Antibody Cocktail. The heatmaps display the PTM levels of Stat3 and MAPK before and after stimulation, as well as which RNA and proteins correlate with those shifts in phosphorylation levels.
Optimize Your Single-Cell Analysis Research
InTraSeq can be a unique and powerful screening tool when starting a project. It enables researchers to understand a specific genetic phenotype by offering an unbiased approach that determines which signaling pathway is perturbed and can help determine where to focus.
InTraSeq can also measure acute cellular perturbations that occur within short periods of time (< 20 minutes) since PTM responses occur before mRNA. These acute responses would otherwise not be observed in RNA alone in single cells and highlight the importance and uniqueness of InTraSeq technology.
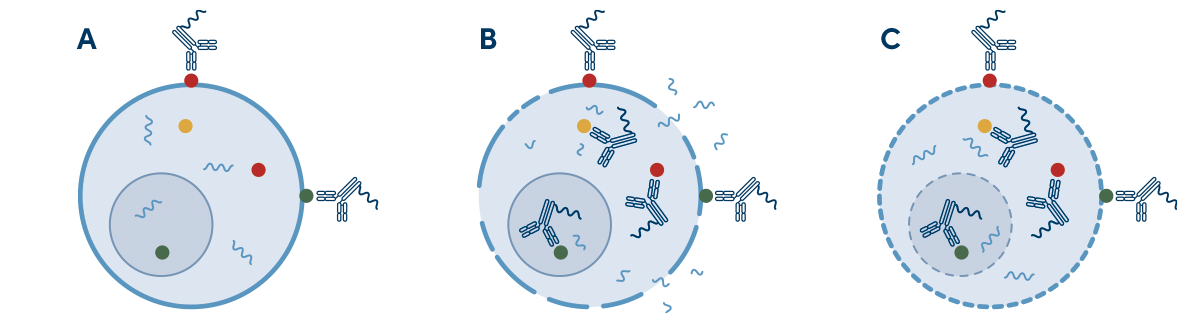
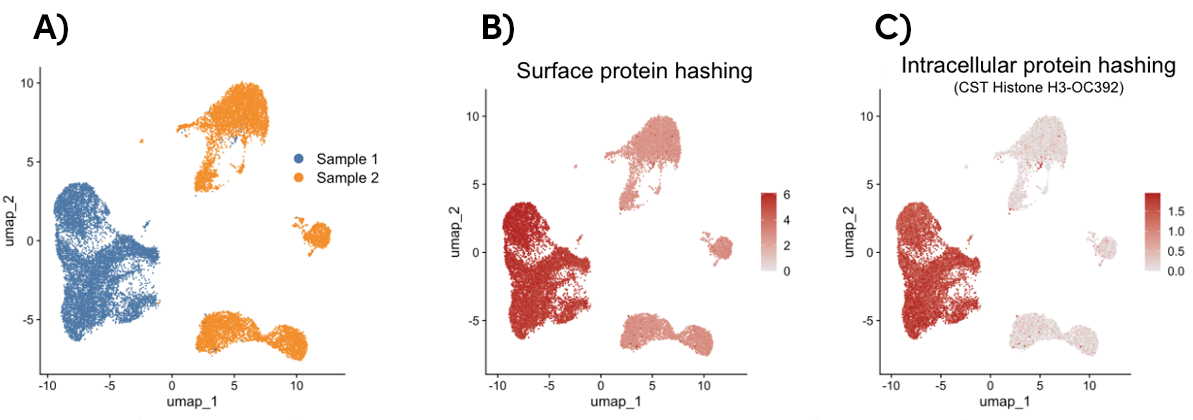
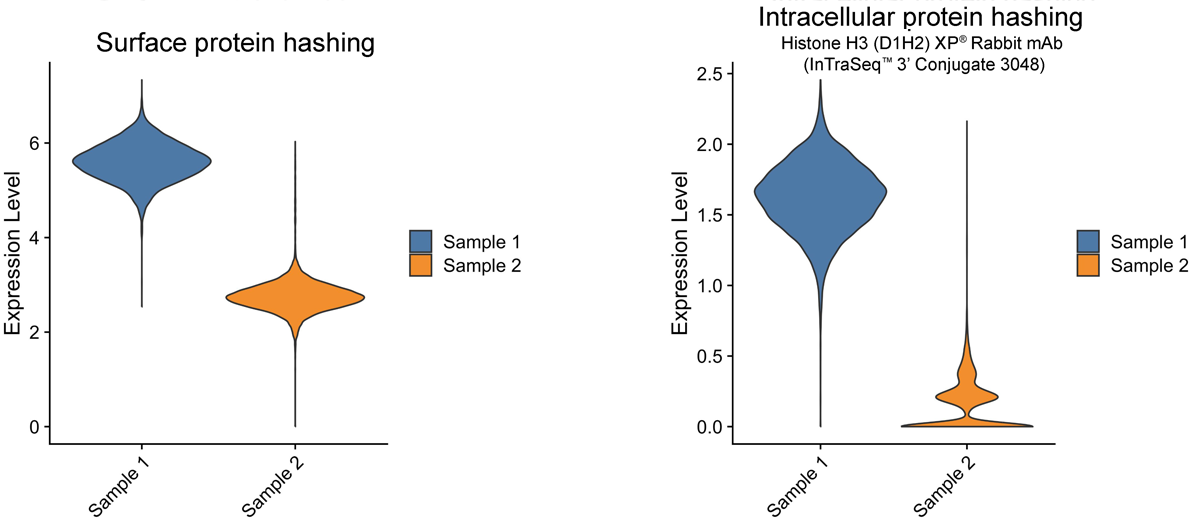
Increase Accuracy with Intracellular Proteins in Sample Multiplexing
In single-cell analysis, sample multiplexing is a technique that uses either surface proteins or intracellular proteins to individually label multiple samples prior to a single-cell experiment. This results in both cost and time savings, as it minimizes the number of single-cell RNA-seq experiments and increases experimental throughput. It also reduces batch variability to generate reproducible data.
Histone H3 is a ubiquitously expressed nuclear protein leveraged in InTraSeq samples to generate a streamlined multiplexing protocol that offers more specificity than surface multiplexing and facilitates the de-multiplexing process, as shown in Figure 1.
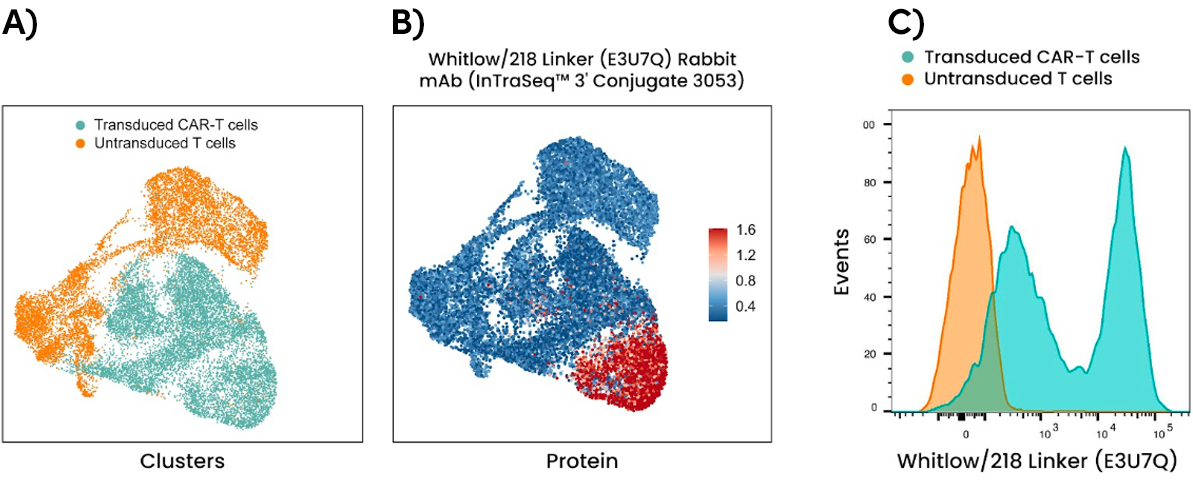
Identify CAR-Expressing Cells with Single-Cell Resolution
Chimeric antigen receptor (CAR) T-cell therapy has shown tremendous promise in leveraging the body’s immune system to direct an antitumor response. Highly specific anti-CAR linker antibodies enable the monitoring of surface expression for nearly any scFv-based CAR, which often contain either a repeating G4S or Whitlow/218 linker sequence.
The Whitlow/218 Linker (E3U7Q) Rabbit mAb (InTraSeq™ 3’ Conjugates 3053) for single-cell analysis identifies CAR-expressing cells at the single-cell resolution and simultaneously reveals multiple signaling pathways to uncover underlying biology.
Visit CAR-Engineered Cell Characterization Solutions to learn more about CST CAR linker antibodies, kits, and services.
Trust Your Results with the CST-Validated InTraSeq™ 3' Conjugate Antibody Cocktail
CST scientists rigorously validate and optimize all InTraSeq antibodies and reagents to provide consistent, high-quality results. The InTraSeq™ 3' Conjugate Antibody Cocktail is formulated such that all the included antibodies and reagents are at optimal concentrations and ready to use in your experiments. InTraSeq removes the guesswork and enables scientists to collect trustworthy data without the need for time-consuming optimization experiments.
InTraSeq Product Offerings for Single Cell-Analysis
Find the right solution for your workflow below.
InTraSeq™ 3' Conjugate Antibody Cocktails
Each of the InTraSeq™ 3' Conjugate Antibody Cocktails contains a mixture of primary antibodies against proteins found in popular signaling pathways. Specially formulated so that you can easily mix and match to create a larger cocktail of your choice. All of these cocktails are reactive toward both human and mouse samples.
InTraSeq™ 3’ Conjugate Antibody Cocktail 1: 31 Primary Antibodies
InTraSeq™ 3’ Conjugate Antibody Cocktail 2: 32 Primary Antibodies
InTraSeq™ 3’ Conjugate Antibody Cocktail 3: 32 Primary Antibodies
A buffer kit specially formulated for a successful 3' single-cell experiment, containing the necessary reagents to ensure robust mRNA and protein signal.
There are seven human-reactive individual conjugates, CAR Linkers, and Histone (H3) for multiplexing are not included in InTraSeq Antibody Cocktails but are available for purchase separately. All InTraSeq Antibody Cocktail targets can also be purchased individually.
Product # | Surface Targets | Species Reactivity | Included in Cocktail? |
|---|---|---|---|
| #72489 | CD3 (UCHT1) Mouse mAb (InTraSeq™ 3' Conjugate 3028) | H | No |
| #87589 | CD4 (RPA-T4) Mouse mAb (InTraSeq™ 3' Conjugate 3029) | H | No |
| #25292 | CD8α (SK1) Mouse mAb (InTraSeq™ 3' Conjugate 3030) | H | No |
| #76014 | CD19 (Intracellular Domain) (D4V4B) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3010) | H, M | Yes |
| #29105 | CD68 (D4B9C) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3012) | H | No |
| #17655 | NCAM1 (CD56) (E7X9M) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3025) | H, M | Yes |
| #57412 | T-bet/TBX21 (D6N8B) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3009) | H | No |
Intracellular Targets (non-PTM) | Species Reactivity | Included in Cocktail? | |
|---|---|---|---|
| #61429 | Aiolos (D1C1E) Rabbit mAb #15103 (InTraSeq™ 3' Conjugate 3027) | H, M | Yes |
| #45300 | FoxO1 (C29H4) Rabbit mAb (InTraSeq™ 3' Conjugate 3037) | H, M | Yes |
| #27907 | GAPDH (14C10) Rabbit mAb (InTraSeq™ 3' Conjugate 3007) | H, M | Yes |
| #82746 | Iba1/AIF-1 (E4O4W) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3003) | H, M | Yes |
| #42531 | Ikaros (D6N9Y) Rabbit mAb #14859 (InTraSeq™ 3' Conjugate 3026) | H, M | Yes |
| #53722 | NFAT1 (D43B1) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3032) | H, M | Yes |
| #97056 | S100A9 (D5O6O) Rabbit mAb (InTraSeq™ 3' Conjugate 3004) | H | No |
| #44070 | TCF1/TCF7 (C63D9) Rabbit mAb (InTraSeq™ 3' Conjugate 3013) | H, M | Yes |
| #96524 | Vimentin (D21H3) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3011) | H, M | Yes |
| PTM-related Targets | Species Reactivity | Included in Cocktail? | |
|---|---|---|---|
| #45003 | Phospho-4E-BP1 (Thr37/46) (236B4) Rabbit mAb (InTraSeq™ 3' Conjugate 3021) | H, M | Yes |
| #35123 | Akt (pan) (C67E7) Rabbit mAb (InTraSeq™ 3' Conjugate 3018) | H, M | Yes |
| #53639 | Phospho-Akt (Ser473) (D9E) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3014) | H, M | Yes |
| #41320 | CREB (48H2) Rabbit mAb (InTraSeq™ 3' Conjugate 3008) | H, M | Yes |
| #95580 | Phospho-CREB (Ser133) (87G3) Rabbit mAb (InTraSeq™ 3' Conjugate 3006) | H, M | Yes |
| #25504 | Glucocorticoid Receptor (D6H2L) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3035) | H, M | Yes |
| #25665 | Phospho-Glucocorticoid Receptor (Ser226) (D9D3V) Rabbit mAb (InTraSeq™ 3' Conjugate 3036) | H, M | Yes |
| #27234 | Acetyl-Histone H3 (Lys27) (D5E4) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3020) | H, M | Yes |
| #92612 | Phospho-Histone H3 (Ser10) (D2C8) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3024) | H, M | Yes |
| #53207 | Tri-Methyl-Histone H3 (Lys27) (C36B11) Rabbit mAb (InTraSeq™ 3' Conjugate 3017) | H, M | Yes |
| #76695 | NF-κB p65 (D14E12) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3033) | H, M | Yes |
| #97410 | Phospho-NF-κB p65 (Ser536) (93H1) Rabbit mAb (InTraSeq™ 3' Conjugate 3034) | H, M | Yes |
| #79941 | p44/42 MAPK (Erk1/2) (137F5) Rabbit mAb (InTraSeq™ 3' Conjugate 3016) | H, M | Yes |
| #76939 | Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (197G2) Rabbit mAb (InTraSeq™ 3' Conjugate 3023) | H, M | Yes |
| #38889 | S6 Ribosomal Protein (54D2) Mouse mAb (InTraSeq™ 3' Conjugate 3005) | H, M | Yes |
| #92243 | Phospho-S6 Ribosomal Protein (Ser235/236) (D57.2.2E) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3019) | H, M | Yes |
| #41599 | Stat1 (D1K9Y) Rabbit mAb (InTraSeq™ 3' Conjugate 3038) | H, M | Yes |
| #82887 | Phospho-Stat1 (Ser727) (D3B7) Rabbit mAb (InTraSeq™ 3' Conjugate 3039) | H, M | Yes |
| #68474 | Stat3 (124H6) Mouse mAb (InTraSeq™ 3' Conjugate 3022) | H, M | Yes |
| #69098 | Phospho-Stat3 (Tyr705) (D3A7) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3015) | H, M | Yes |
| #34774 | Phospho-Stat3 (Ser727) (D4X3C) Rabbit mAb (InTraSeq™ 3' Conjugate 3031) | H, M | Yes |
| Isotype Control | Species Reactivity | Included in Cocktail? | |
|---|---|---|---|
| #59605 | Mouse (G3A1) mAb IgG1 Isotype Control (InTraSeq™ 3' Conjugate 3001) | H | No |
| Multiplexing Reagents | Species Reactivity | Included in Cocktail? | |
|---|---|---|---|
| #52488 | Histone H3 (D1H2) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3048) | H, M | No |
| #77931 | Histone H3 (D1H2) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3049) | H, M | No |
| #98269 | Histone H3 (D1H2) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3050) | H, M | No |
| #83332 | Histone H3 (D1H2) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3051) | H, M | No |
| #32730 | Histone H3 (D1H2) XP® Rabbit mAb (InTraSeq™ 3' Conjugate 3052) | H, M | No |
| CAR Linkers | Species Reactivity | Included in Cocktail? | |
|---|---|---|---|
| #50265 | Whitlow/218 Linker (E3U7Q) Rabbit mAb (InTraSeq™ 3' Conjugate 3053) | All | No |